Revision 1
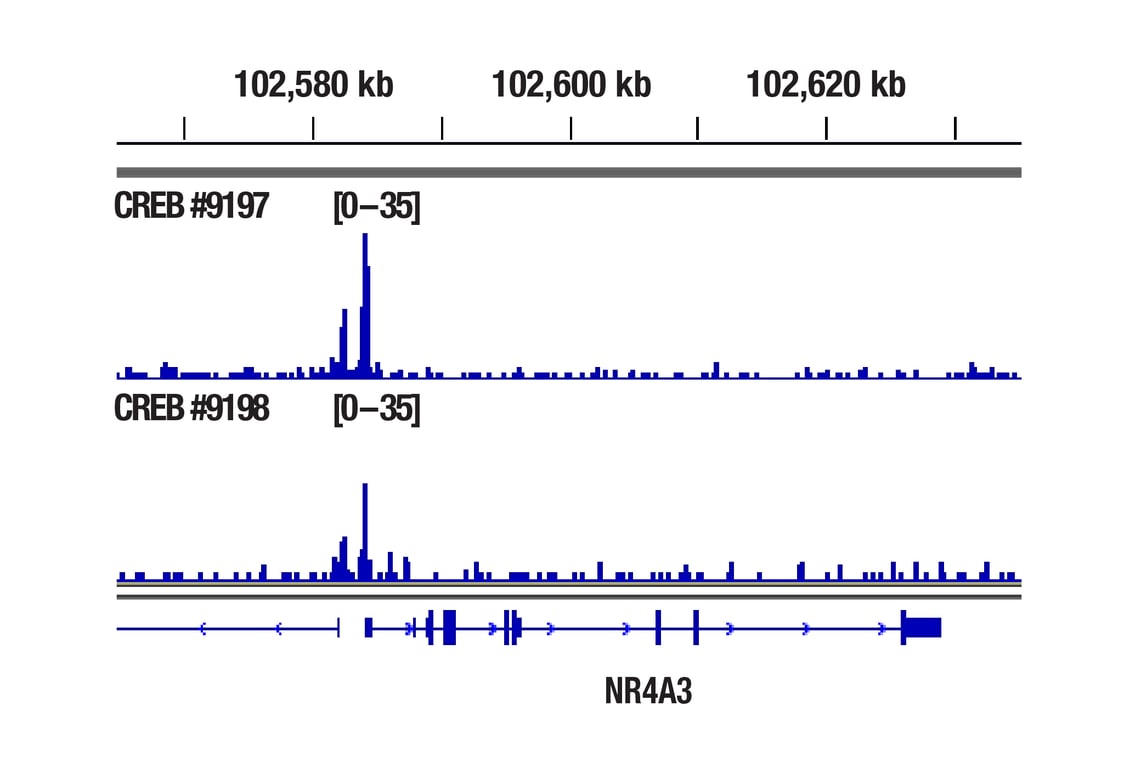
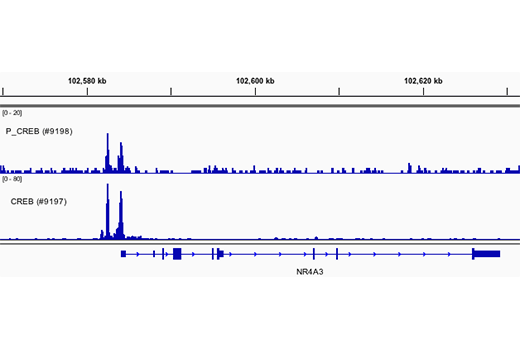
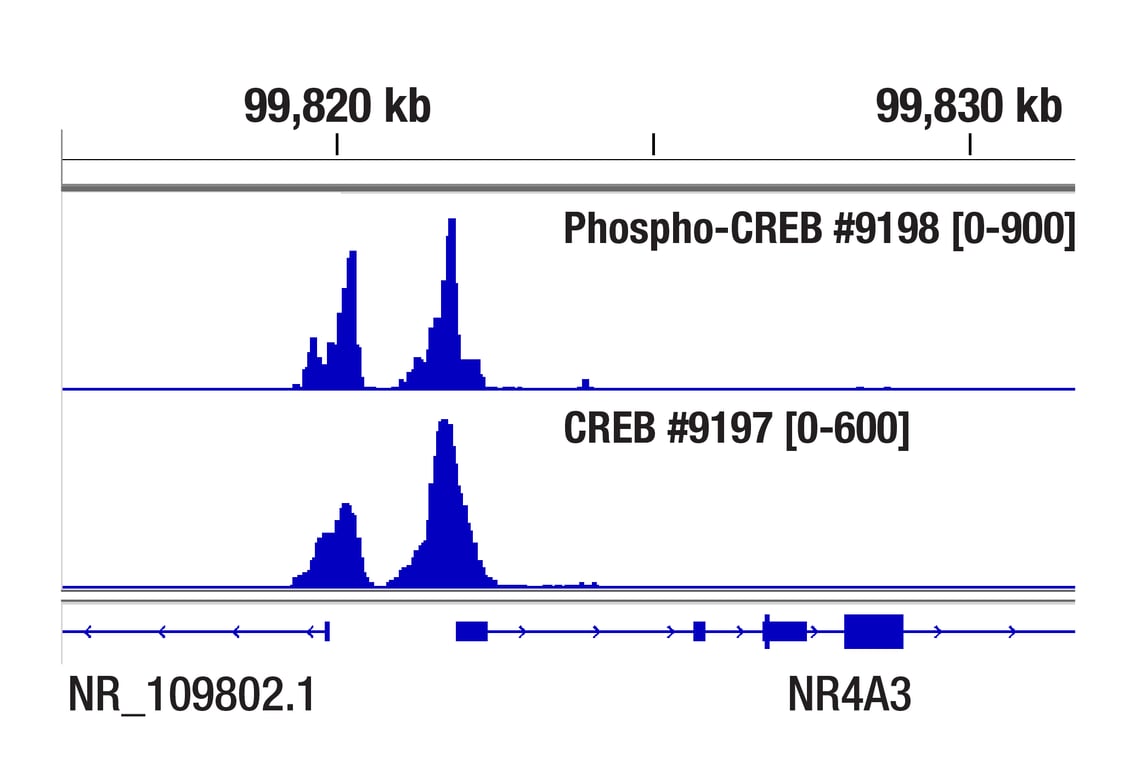
CUT&RUN was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either CREB (48H2) Rabbit mAb or Phospho-CREB (Ser133) (87G3) Rabbit mAb #9198, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795. The figure shows binding across NR4A3 gene.
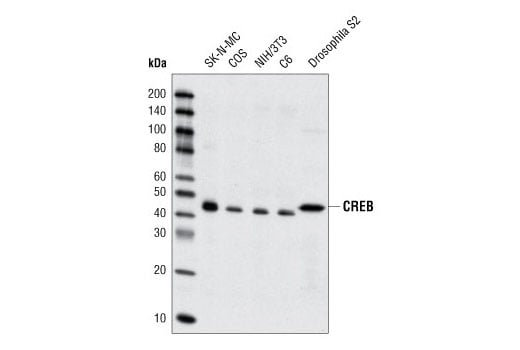
Western Blot analysis of extracts from SK-N-MC, COS, NIH/3T3, C6 and Drosophila S2 cells, using CREB (48H2) Rabbit mAb.
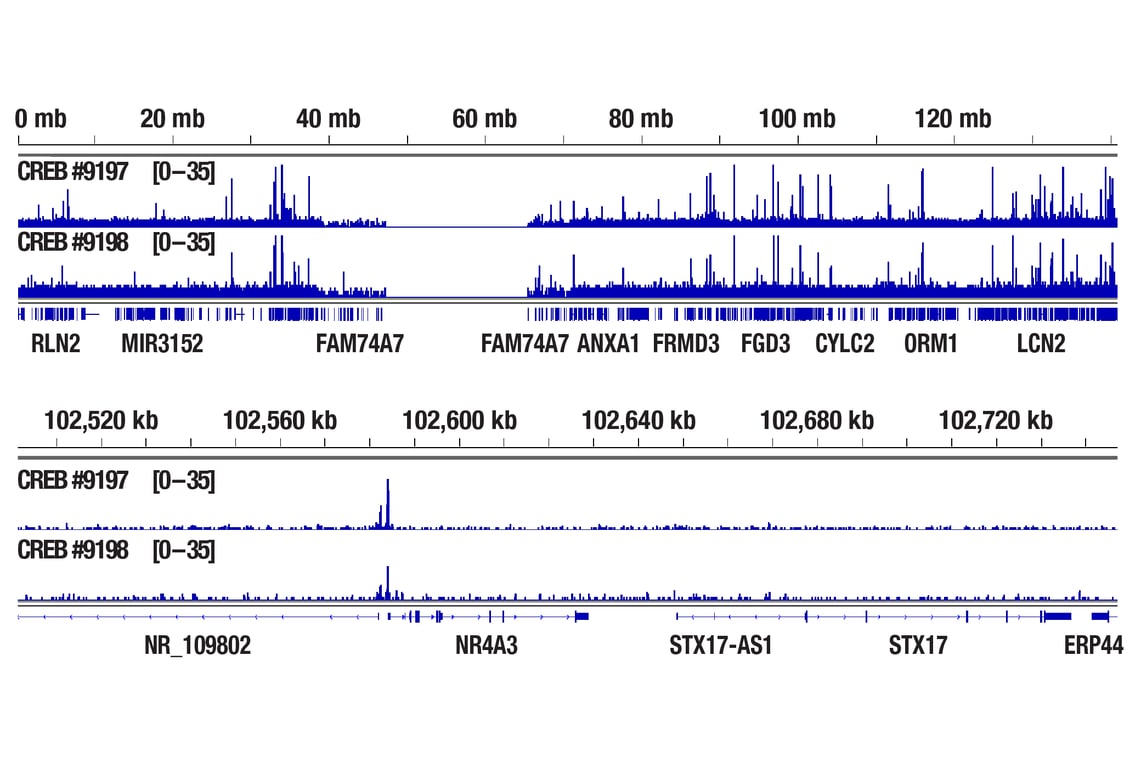
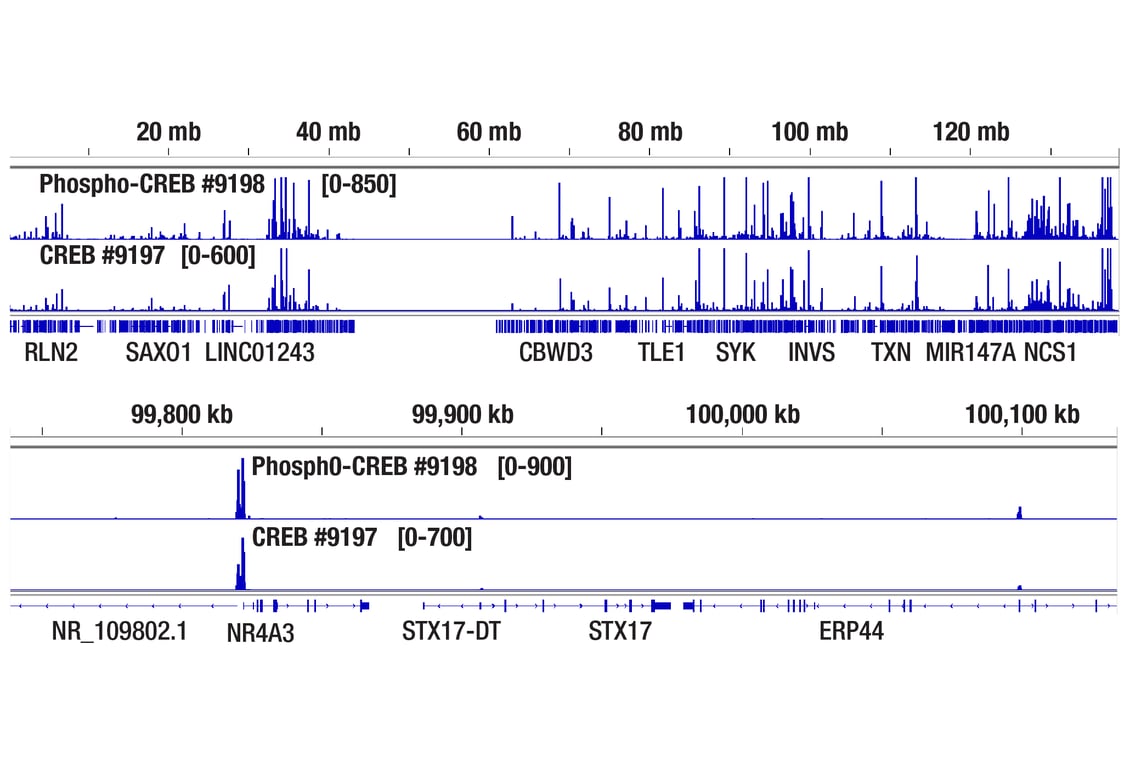
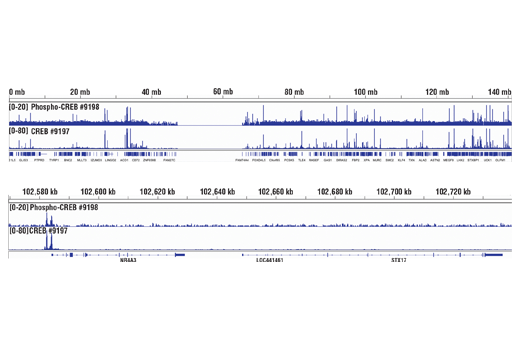
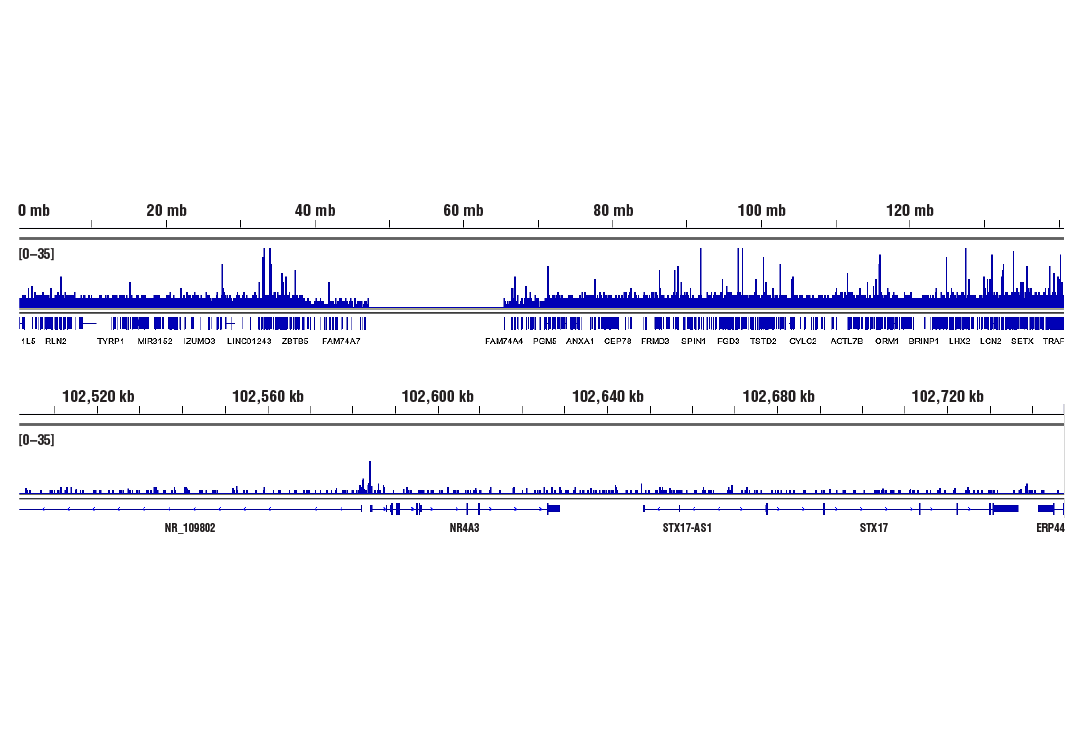
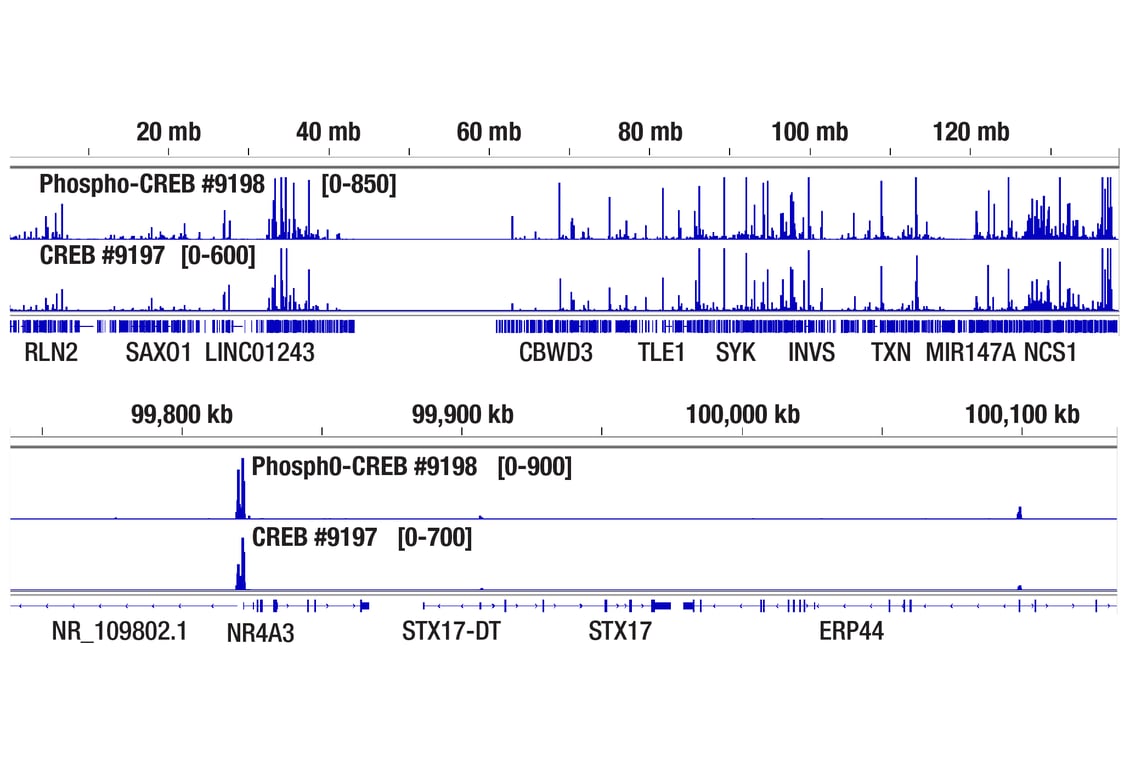
CUT&RUN was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either CREB (48H2) Rabbit mAb or Phospho-CREB (Ser133) (87G3) Rabbit mAb #9198, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using DNA Library Prep Kit for Illumina Systems (ChIP-seq, CUT&RUN) #56795. The figures show binding across chromosome 9 (upper), including NR4A3 (lower) gene.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
CUT&Tag was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either CREB (48H2) Rabbit mAb or Phospho-CREB (Ser133) (87G3) Rabbit mAb #9198, using CUT&Tag Assay Kit #77552. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across NR4A3, a known target gene of both CREB and Phospho-CREB (see additional figure containing ChIP-qPCR data).
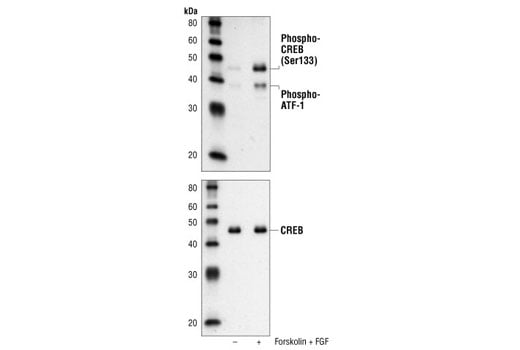
Western blot analysis of extracts from SK-N-MC cells, untreated or forskolin- and FGF-treated, using Phospho-CREB (Ser133) (87G3) Rabbit mAb (upper) or CREB (48H2) Rabbit mAb #9197 (lower).
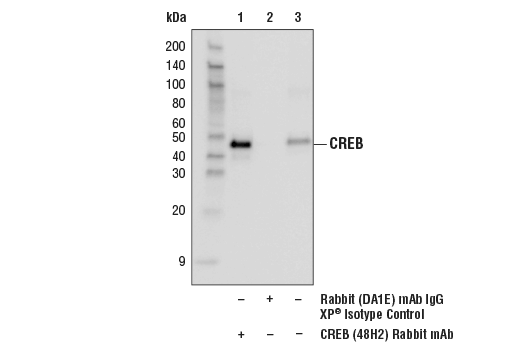
Immunoprecipitation of CREB from SK-N-MC extracts. Lane 1 is CREB (48H2) Rabbit mAb, lane 2 is Rabbit (DA1E) mAb IgG XP® Isotype Control #3900, and lane 3 is 10% input. Western blot analysis was perfomed using CREB (86B10) Mouse mAb #9104. Anti-mouse IgG, HRP-linked Antibody #7076 was used as a secondary antibody.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
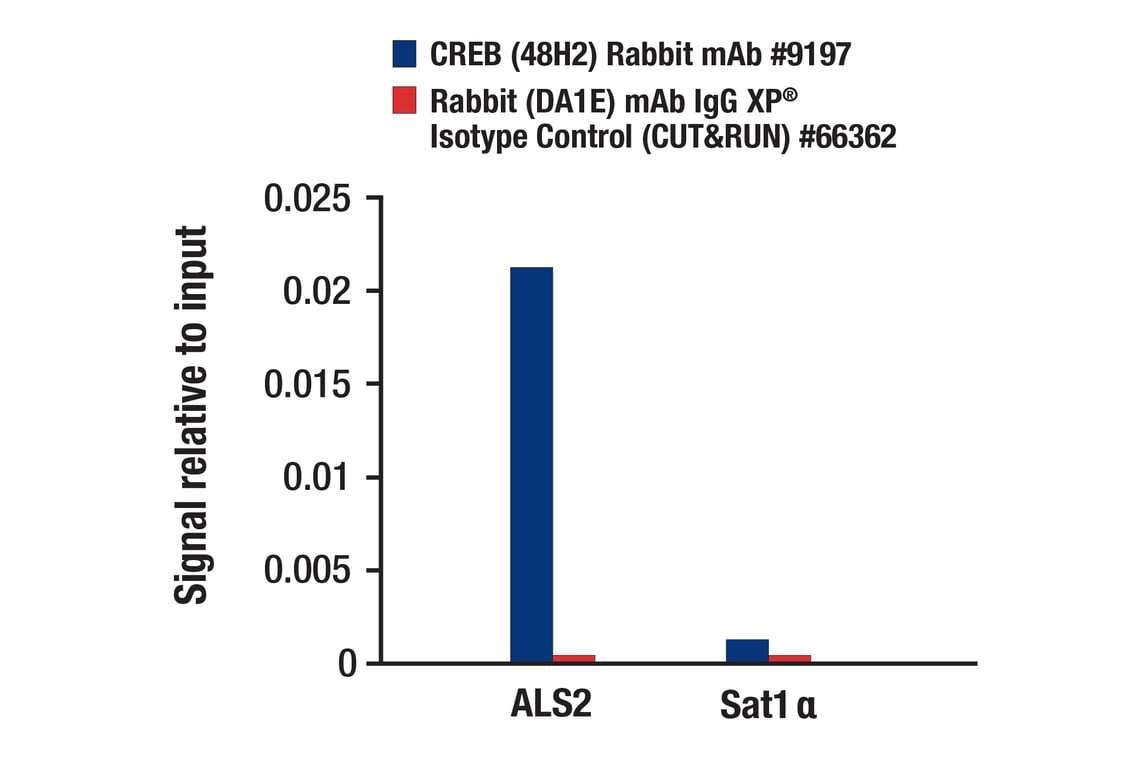
CUT&RUN was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either CREB (48H2) Rabbit mAb or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using CUT&RUN Assay Kit #86652. The enriched DNA was quantified by real-time PCR using human ALS2 exon 1 primers and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
CUT&Tag was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either CREB (48H2) Rabbit mAb or Phospho-CREB (Ser133) (87G3) Rabbit mAb #9198, using CUT&Tag Assay Kit #77552. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across chromosome 9 (upper), including NR4A3 (lower), a known target gene of both CREB and Phospho-CREB (see additional figure containing ChIP-qPCR data).
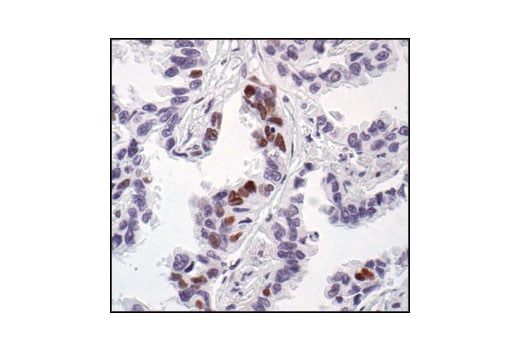
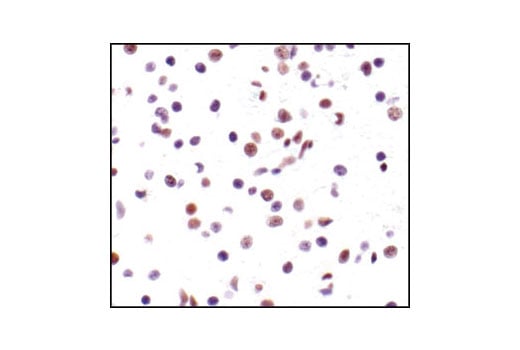
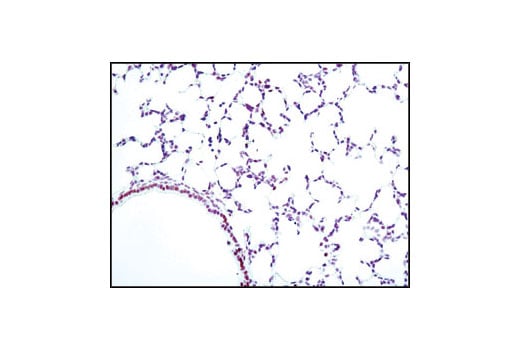
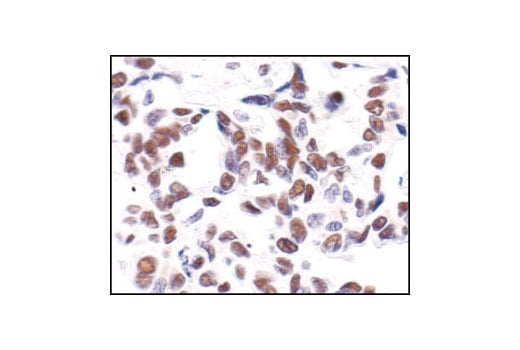
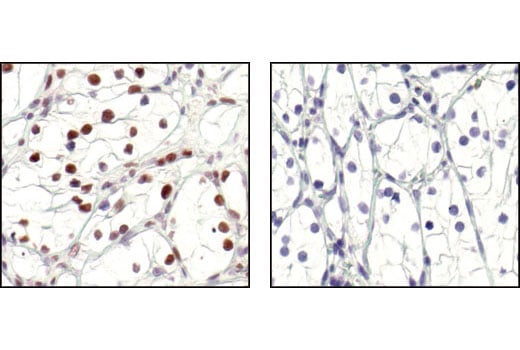
Immunohistochemical analysis of paraffin-embedded human lung carcinoma, showing nuclear staining, using Phospho-CREB (Ser133) (87G3) Rabbit mAb.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
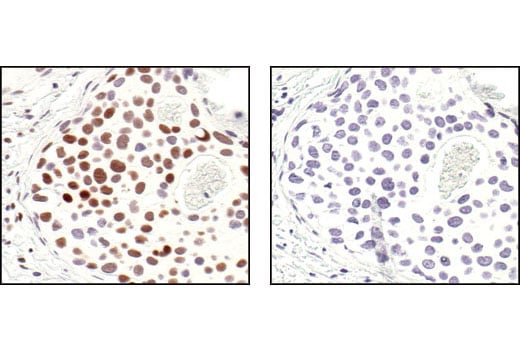
Immunohistochemical analysis of paraffin-embedded human astrocytoma, using CREB (48H2) Rabbit mAb.
Immunohistochemical analysis of paraffin-embedded mouse lung using Phospho-CREB (Ser133) (87G3) Rabbit mAb.
Immunohistochemical analysis of paraffin-embedded human lung carcinoma, using CREB (48H2) Rabbit mAb.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
Immunohistochemical analysis of paraffin-embedded human breast carcinoma, using Phospho-CREB (Ser133) (87G3) Rabbit mAb in the presence of control peptide (left) or Phospho-CREB (Ser133) Blocking Peptide #1090 (right).
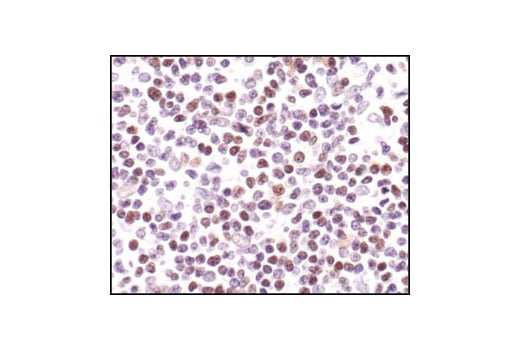
Immunohistochemical analysis of paraffin-embedded human Non-Hodgkin's lymphoma, using CREB (48H2) Rabbit mAb.
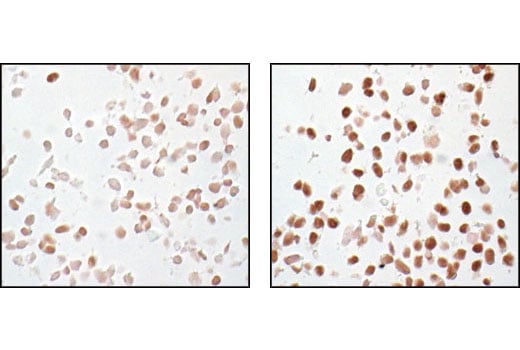
Immunohistochemical analysis of paraffin-embedded SK-N-MC cells, untreated (left) or IBMX- and forskolin-treated (right), showing induced nuclear staining, using Phospho-CREB (Ser133) (87G3) Rabbit mAb.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
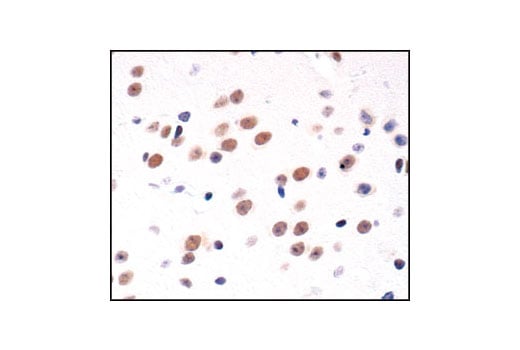
Immunohistochemical analysis of paraffin-embedded mouse brain, using CREB (48H2) Rabbit mAb.
Immunohistochemical analysis of paraffin-embedded human renal cell carcinoma, untreated (left) or lambda phosphatase-treated (right), using Phospho-CREB (Ser133) (87G3) Rabbit mAb.
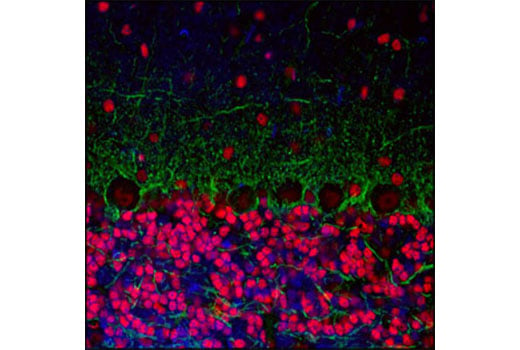
Confocal immunofluorescent analysis of mouse cerebellum labeled with CREB (48H2) Rabbit mAb (red) and Neurofilament-L (DA2) Mouse mAb #2835 (green). Blue pseudocolor =DRAQ5® #4084 (fluorescent DNA dye).
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
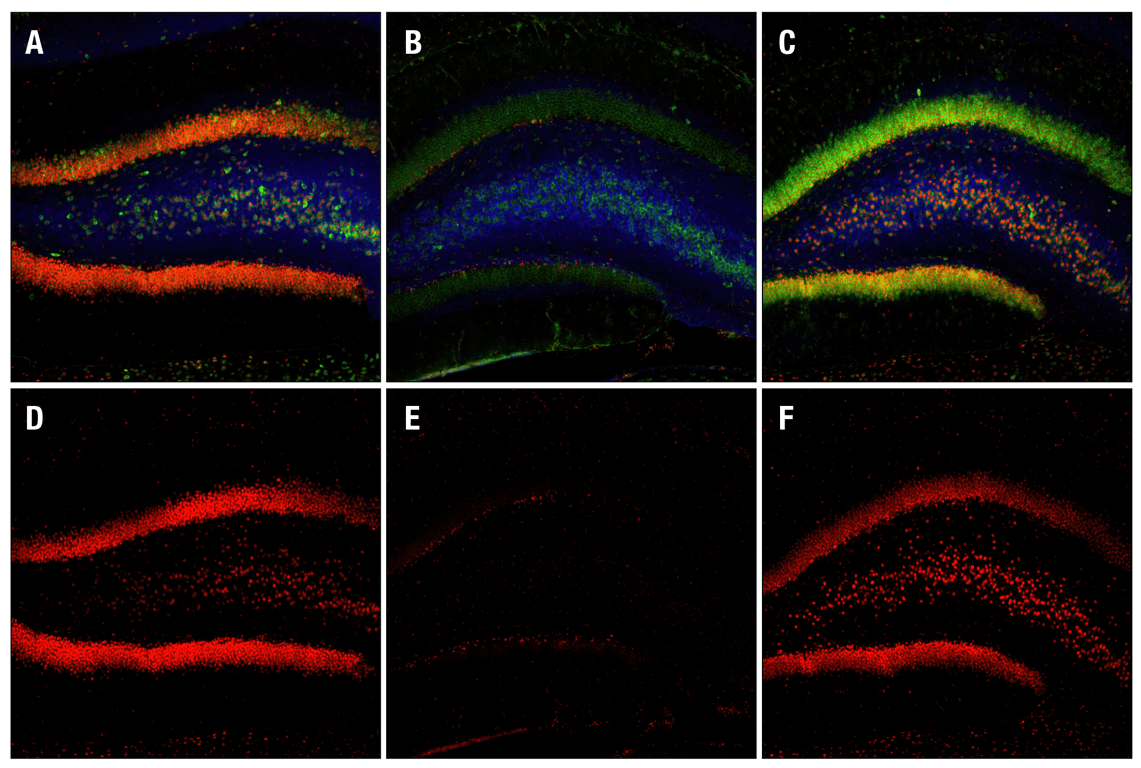
Confocal immunofluorescent images of rat dentate gyrus, either sham-operated (left) or 15 min ischemia followed by 30 min (center) and 4 h (right) reperfusion, labeled with Phospho-CREB (Ser133) (87G3) Rabbit mAb (red), Neurofilament-L (DA2) Mouse mAb #2835 (blue) and Phospho-S6 Ribosomal Protein (Ser235/236) (2F9) Rabbit mAb (Alexa Fluor® 488 Conjugate) #4854.
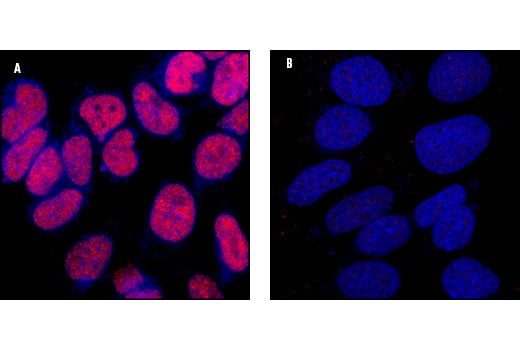
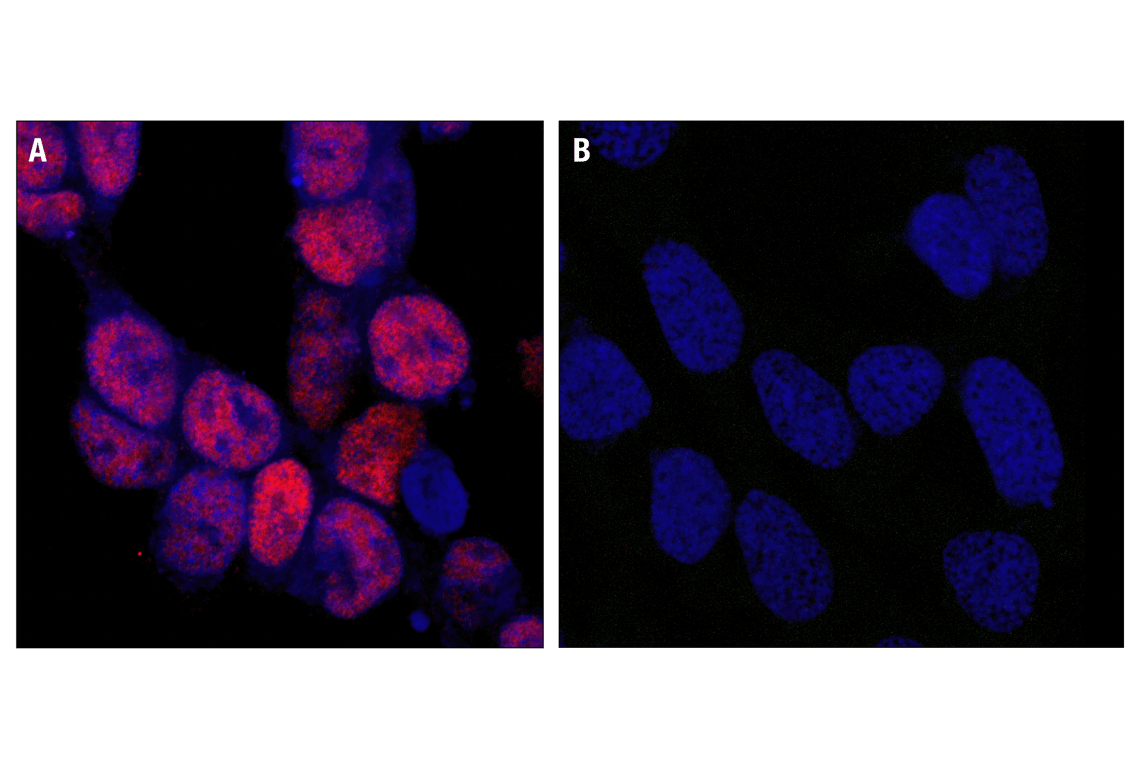
Confocal immunofluorescent analysis of SK-N-MC cells showing nuclear stain with CREB (48H2) Rabbit mAb (A, red) compared to an isotype control (B). Blue pseudocolor =DRAQ5® (fluorescent DNA dye).
Confocal microscopic images of SK-N-MC cells showing nuclear stain after 25 minute treatment with Forskolin and IBMX using Phospho-CREB (Ser133) (87G3) Rabbit mAb (left, red) compared to untreated cells (right). Blue pseudocolor = DRAQ5® #4084 (fluorescent DNA dye).
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
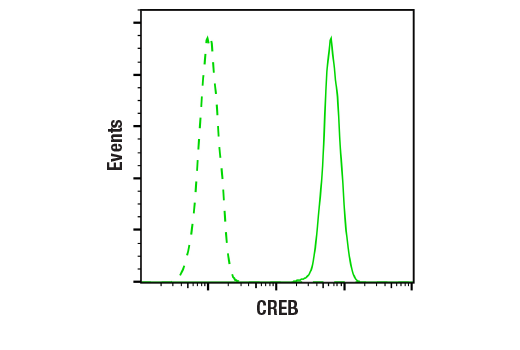
Flow cytometric analysis of Jurkat cells using CREB (48H2) Rabbit mAb (solid line) compared to concentration-matched Rabbit (DA1E) mAb IgG XP® Isotype Control #3900 (dashed line). Anti-rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) #4412 was used as a secondary antibody.
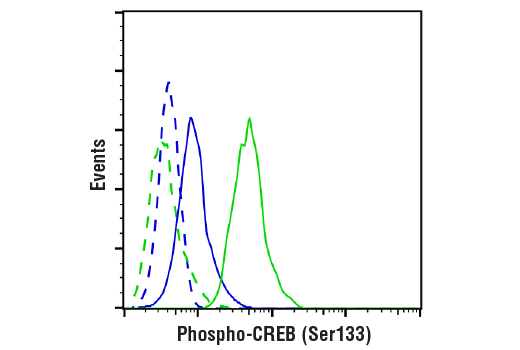
Flow cytometric analysis of Jurkat cells, untreated (blue) or treated with TPA #4174 (500nM) and Ionomycin #9995 (1uM,4hrs;green) using Phospho-CREB (Ser133) (87G3) Rabbit mAb (solid lines) or concentration-matched Rabbit (DA1E) mAb IgG XP® Isotype Control #3900 (dashed lines). Anti-rabbit IgG (H+L), F(ab')2 Fragment (Alexa Fluor® 488 Conjugate) #4412 was used as a secondary antibody.
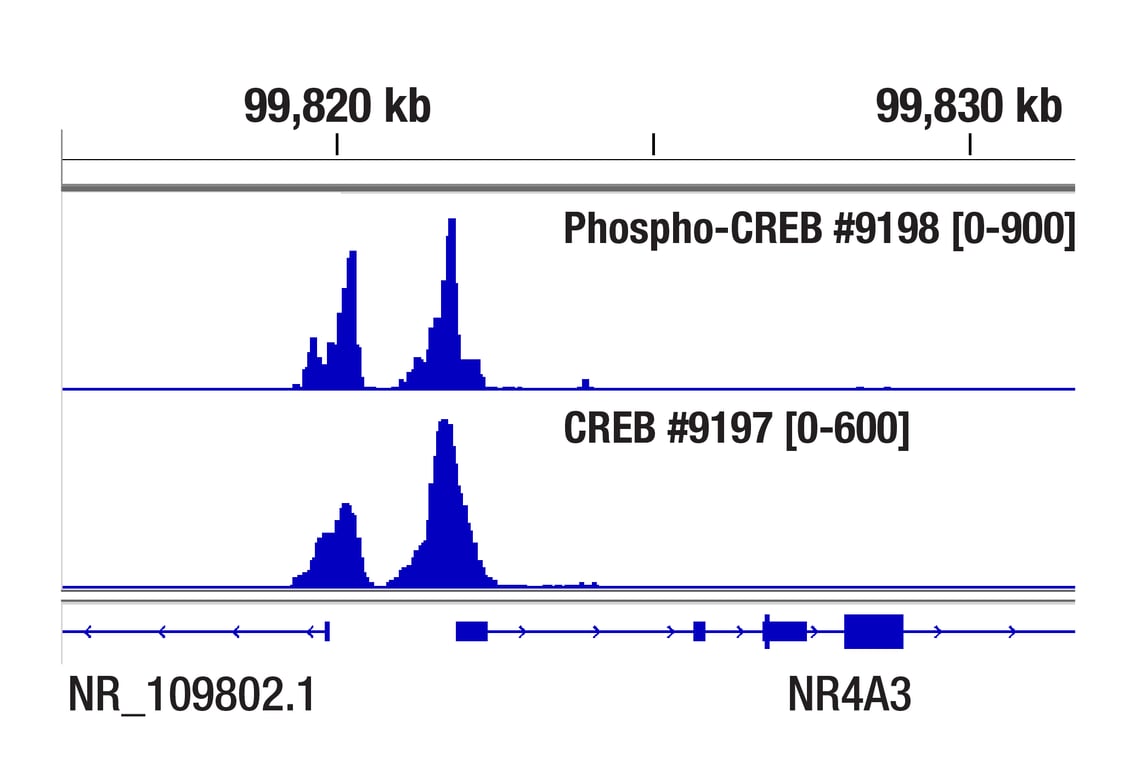
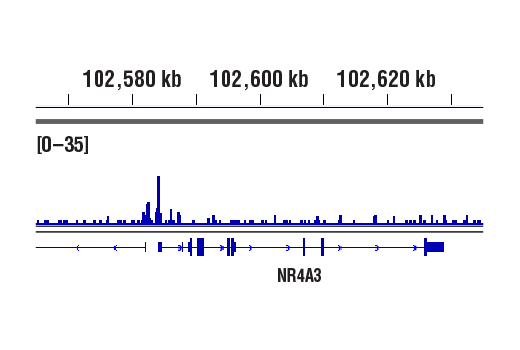
Chromatin immunoprecipitations were performed with cross-linked chromatin from 293 cells treated with Forskolin #3828 (30 μM) for 1h and either Phospho-CREB (Ser133) (87G3) Rabbit mAb or CREB (48H2) Rabbit mAb (#9197), using SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The figure shows binding across NR4A3, a known target gene of both Phospho-CREB and CREB (see additional figure containing ChIP-qPCR data).
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
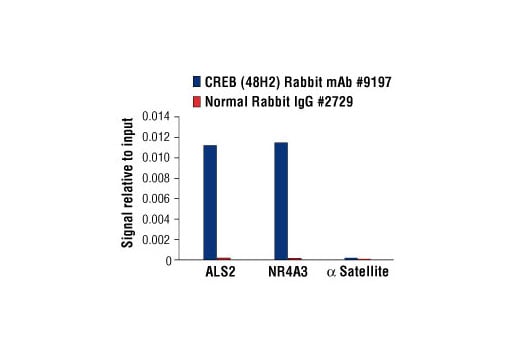
Chromatin immunoprecipitations were performed with cross-linked chromatin from 293 cells, treated with Forskolin #3828 (30 μM) for 1h and either 10 μl of CREB (48H2) Rabbit mAb or 2 μl of Normal Rabbit IgG #2729, using SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003. The enriched DNA was quantified by real-time PCR using human ALS2 exon 1 primers, SimpleChIP® Human NR4A3 Promoter Primers #4829, and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
Chromatin immunoprecipitations were performed with cross-linked chromatin from 293 cells treated with Forskolin #3828 (30 μM) for 1h and either Phospho-CREB (Ser133) (87G3) Rabbit mAb or CREB (48H2) Rabbit mAb (#9197), using SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9005. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The figure shows binding across chromosome 9 (upper), including NR4A3 (lower), a known target gene of both Phospho-CREB and CREB (see additional figure containing ChIP-qPCR data).
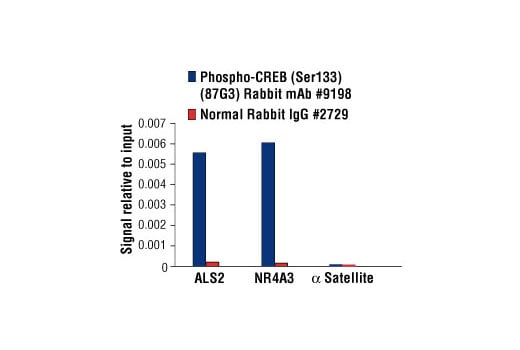
Chromatin immunoprecipitations were performed with cross-linked chromatin from 293 cells treated with Forskolin #3828 (30 μM) for 1h and either Phospho-CREB (Ser133) (87G3) Rabbit mAb or Normal Rabbit IgG #2729 using SimpleChIP® Enzymatic Chromatin IP Kit (Magnetic Beads) #9003. The enriched DNA was quantified by real-time PCR using human ALS2 exon 1 primers, SimpleChIP® Human NR4A3 Promoter Primers #4829, and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
CUT&RUN was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and Phospho-CREB (Ser133) (87G3) Rabbit mAb, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® (ChIP-seq, CUT&RUN) #56795. The figure shows binding across NR4A3 gene.
CUT&RUN was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and Phospho-CREB (Ser133) (87G3) Rabbit mAb, using CUT&RUN Assay Kit #86652. DNA Libraries were prepared using SimpleChIP® ChIP-seq DNA Library Prep Kit for Illumina® (ChIP-Seq, CUT&RUN) #56795. The figures show binding across chromosome 9 (upper), including NR4A3 (lower) gene.
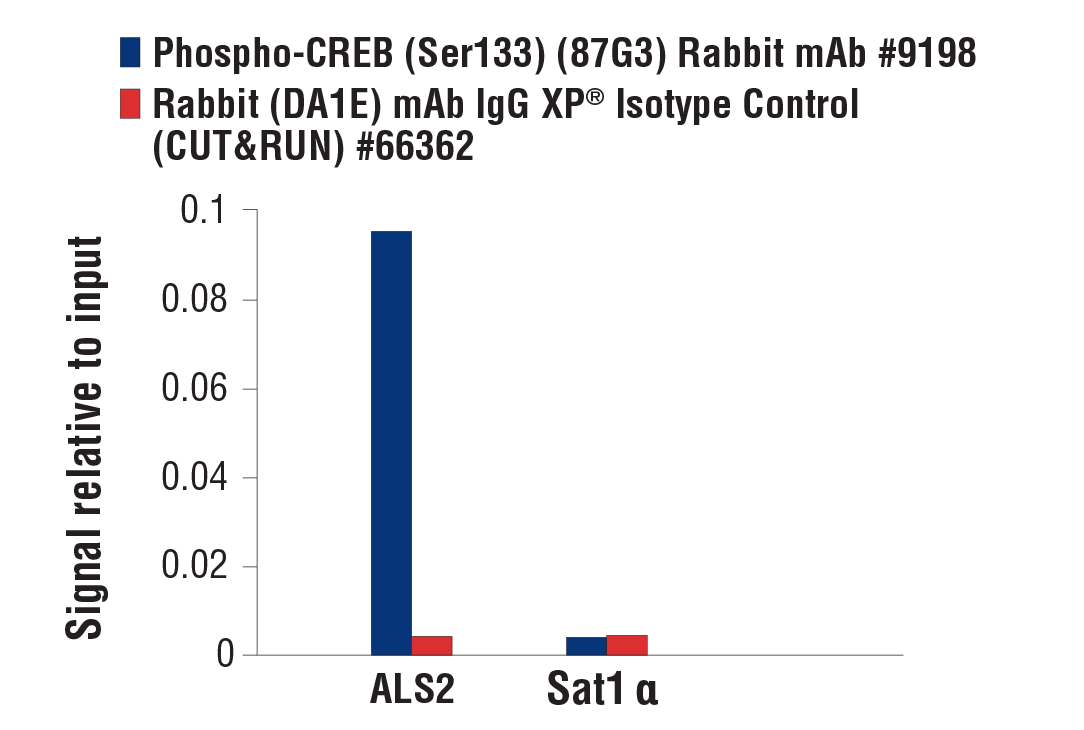
CUT&RUN was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either Phospho-CREB (Ser133) (87G3) Rabbit mAb or Rabbit (DA1E) mAb IgG XP® Isotype Control (CUT&RUN) #66362, using CUT&RUN Assay Kit #86652. The enriched DNA was quantified by real-time PCR using human ALS2 exon 1 primers and SimpleChIP® Human α Satellite Repeat Primers #4486. The amount of immunoprecipitated DNA in each sample is represented as signal relative to the total amount of input chromatin, which is equivalent to one.
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.
Revision 1
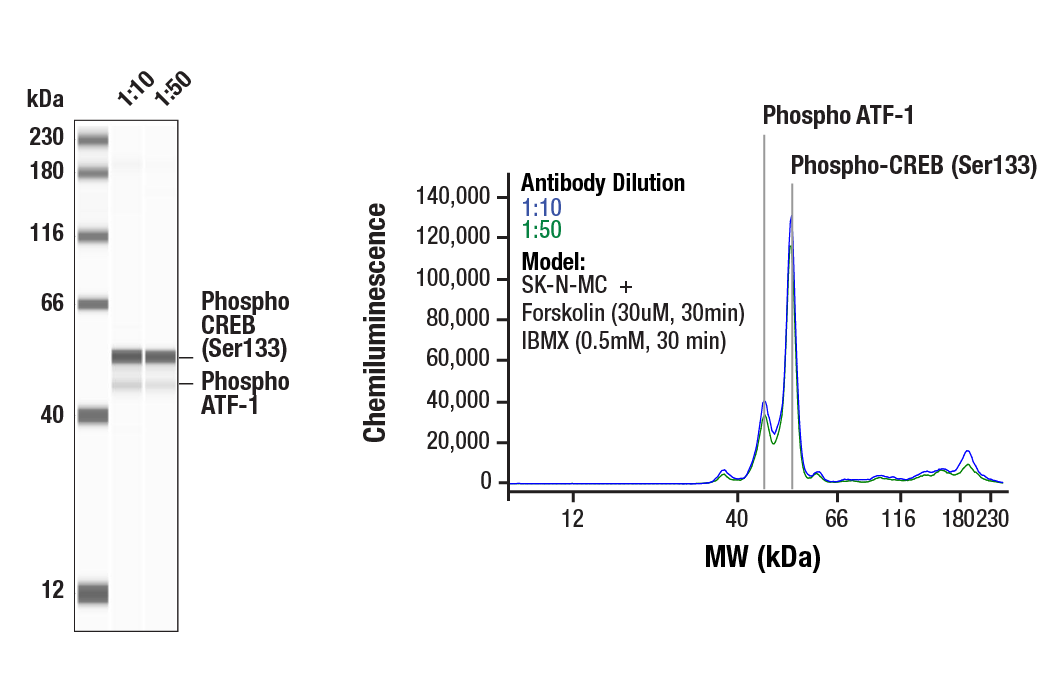
Simple Western™ analysis of lysates (0.1 mg/mL) from SK-N-MC cells treated with Forskolin (30uM, 30min) and IBMX (0.5mM, 30 min) using Phospho-CREB (Ser133) (87G3) Rabbit mAb #9198. The virtual lane view (left) shows the target band (as indicated) at 1:10 and 1:50 dilutions of primary antibody. This antibody also detects the phosphorylated form of the CREB-related protein, ATF-1 (as indicated). The corresponding electropherogram view (right) plots chemiluminescence by molecular weight along the capillary at 1:10 (blue line) and 1:50 (green line) dilutions of primary antibody. This experiment was performed under reducing conditions on the Jess™ Simple Western instrument from ProteinSimple, a BioTechne brand, using the 12-230 kDa separation module.
CUT&Tag was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either Phospho-CREB (Ser133) (87G3) Rabbit mAb or CREB (48H2) Rabbit mAb #9197, using CUT&Tag Assay Kit #77552. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figure shows binding across NR4A3, a known target gene of both Phospho-CREB and CREB (see additional figure containing ChIP-qPCR data).
CUT&Tag was performed with 293 cells treated with Forskolin #3828 (30 μM) for 1h and either Phospho-CREB (Ser133) (87G3) Rabbit mAb or CREB (48H2) Rabbit mAb #9197, using CUT&Tag Assay Kit #77552. DNA libraries were prepared using CUT&Tag Dual Index Primers and PCR Master Mix for Illumina Systems #47415. The figures show binding across chromosome 9 (upper), including NR4A3 (lower), a known target gene of both Phospho-CREB and CREB (see additional figure containing ChIP-qPCR data).
Orders: 877-616-CELL (2355) • [email protected] • Support: 877-678-TECH (8324) • [email protected] •
Web:
cellsignal.com For Research Use Only. Not for Use in Diagnostic Procedures.